
Databases: Bookshelf · CGDB · KDB · OMPDB · Lipidbook | Links | Contact | Intranet | Search
 |
|
Home |
Mission |
Research |
Members |
Positions |
Image Gallery |
Panoramas |
Seminars Databases: Bookshelf · CGDB · KDB · OMPDB · Lipidbook | Links | Contact | Intranet | Search |
Benjamin Hall |
|
Structural Bioinformatics and Computational Biochemistry Unit Dept. of Biochemistry University of Oxford South Parks Road Oxford OX13QU U.K. Telephone: (0)1865-613304 Fax: (0)1865-613238 e-mail: |
 |
Multiscale Modelling and Systems BiologySystems biology is a growing field concerned with the mapping and parametrisation of biological networks to make systems level predictions of the effects of small scale changes. Structural bioinformatics is a related field concerned with the modelling of atomic scale phenomenon in biological molecules, in particular molecular motions. The timescales achieved by such methods preclude comparison with most biological phenomena and current experimental techniques in dynamics. Bridging these two disciplines involves the development and extension of coarse grained methods to advance the timescale achievable and the accurate extraction of parameters from atomistic modelling to build coarse models which may themselves be embedded into network models. To this end I have developed and am developing a wide range of new tools for the analysis, classification and prediction of protein properties. Using these tools I am gaining novel insights into proteins which are well understood and investigating proteins new to the group, which are amenable to embedding in a systems based model. My research is funded by OCISB. I've included some visualisations of my work below, but for those interested there is a larger gallery on my personal webpage |
|
Publications |
|
Hall B.A., Sansom M. S. P. Coarse-Grained MD Simulations and Protein−Protein Interactions: The Cohesin−Dockerin System JCTC 2009, 5(9) 2465 Hall B.A., Kaye S. L., Pang A., Perera R., Biggin P. C. Characterizing Protein States by Normal-Mode Frequencies JACS 2007, 129(37) 11394-11401 Haider S., Hall B.A., Sansom M.S.P. Simulations of a protein translocation pore: SecY. Biochemistry 2006, 45 (43) 13018-24 Grottesi A., Domene C., Hall B., Sansom M.S.P. Conformational Dynamics of M2 Helices in KirBac Channels: Helix Flexibility in Relation to Gating via Molecular Dynamics Simulations Biochemistry 2005, 44 (44) 14586-94 Haider S., Grottesi A., Hall B.A., Ashcroft F.M., Sansom M.S.P. (2005) Conformational Dynamics of the Ligand-Binding Domain of Inward Rectifier K Channels as Revealed by MD Simulations: Towards an Understanding of Kir Channel Gating Biophysical Journal 2005, 88 3310-3320 Barrett C.P., Hall B.A., Noble M.E.M. (2004) Dynamite: A Simple Way to Gain Insight into Protein Motions Acta Crystallographica D 2004, 60 (12-1) 2280-2287 |
|
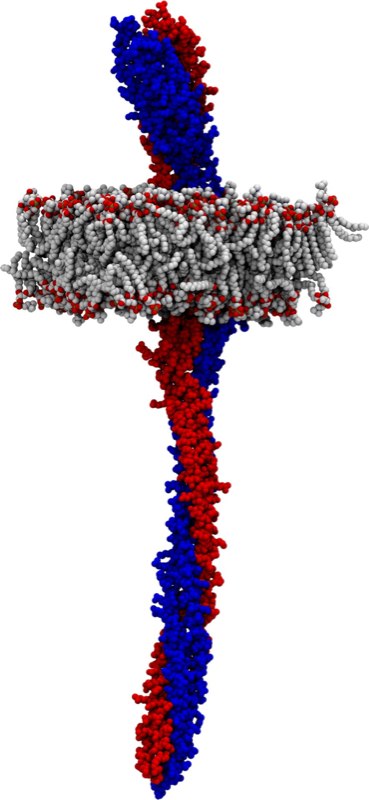
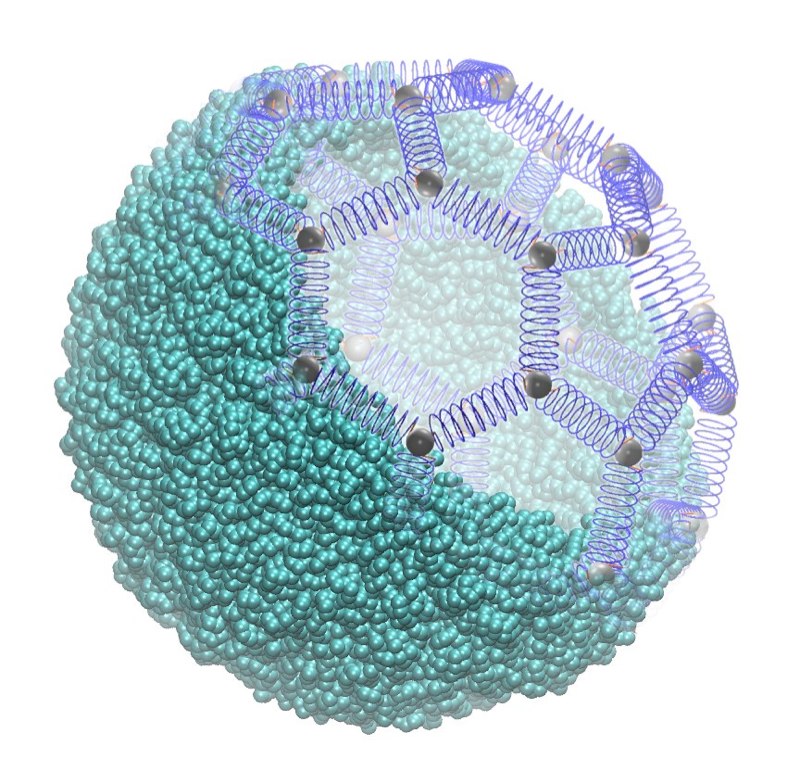
Images |
|

| Last updated 8/09/09 |